The Cellfishing algorithm encodes chemical structures using the Circular Morgan Fingerprints, also known as Extended Chemical Fingerprints (ECFP). We use the Tanimoto coefficient as the molecular similarity metric. The radius used for Circular Morgan Fingerprint is r = 8 (ECFP16). Submit a molecule The compound needs to be submitted in SMILES format.

The molecule is introduced in: “Smile Input”. No salt or disconnected compounds are allowed as input.
The output is a table like:
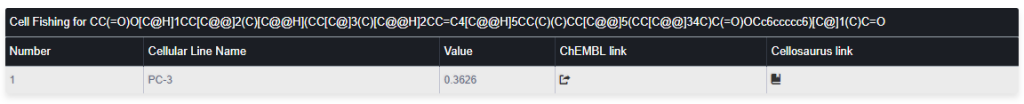
Col 1: Ordering number
Col 2: Cell Name as presented in ChemBL database
Col 3: The Tanimoto similarity average between the query compound and the top three more similar compounds sensible to the presented cell line.
Col 4 and Col 5: Links the cell line information in the ChemBL and Cellosaurus databases respectively. Only cell lines with a similarity higher or equal than 0.35 will be considered as “sensible” to the query compound.
