Set calculation type
The CompScore service operates in two modes. The user has the option of performing either a genetic algorithm (GA) optimization or a rescoring run. The GA optimization finds the combination of scoring functions components maximizing either the Enrichment Factor at a user defined fraction of screened data or BEDROC for a user provided value of the parameter α. For a GA optimization run the user must provide an Data file containing compounds IDs, Classification, Weighting variable and Scoring components value (see the Data file section for details) and setup the GA parameters (see the GA setup section for details). The output of a GA optimization run will be a log file containing details of the calculation and a txt file containing a sorted list, best to worst, of compounds IDs with their aggregated scores (see the Output files section).
For rescoring the user has the options of using a log file from a previous run or to apply the general consensus scoring schemes available for different primary docking programs. The type of rescoring run is selected from the drop down menu. If a custom rescoring is requested, a log file from a previous CompScore run and a Data file must be provided as inputs to apply the found consensus scoring solution to the latter. For the general CompScore model only the Data file is required. The output of a Rescoring run is a txt file containing a sorted list, best to worst, of compounds IDs with their aggregated scores (see the Output files section). This selection of the Calculation Type is made with the radio buttons below and the selection of one of them is mandatory.
Weighted scores selection
For both Rescoring and GA Optimization calculation types the user has the option of including the weighted scores. Weighted scores are internally computed by the algorithm and added to the loaded data. The variable for weighting the scores must be located in the third column of the input data file (see next section for Input data file format). The newly created variables will be identified by the original score ID plus the ‘_W’ suffix. For example, if the ‘Score1’ variable is present in the input data, the newly created variable will be set to ‘Score1_W’. Note that if a Rescoring calculation is requested using weighted scores, a previous optimization run using weighted scores must be previously performed. The user has the option of activating the use of weighted scores by means of the below check box.

Data file
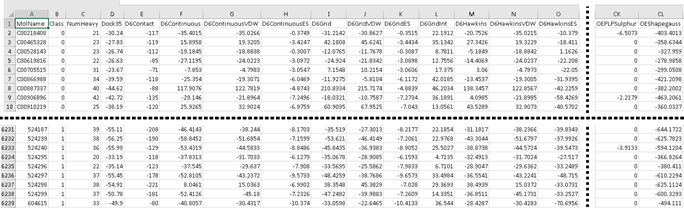
In case a GA optimization run is requested, only the Data input file is required. This file must have the following format:
First row is considered as data heading, i.e variables names.
- Column 1: The ID of each compound.
- Column 2: The classification of the compounds in either ligand of decoy. The allowed values in this column are 1 for ligands and 0 for decoys in case a GA optimization is selected as Calculation type. If a Rescoring calculation is requested, this column can contain any integer data since it won’t be considered during calculations.
- Column 3: The value of the weighting criterion, e.g. number of heavy atoms. It contains numeric values and can have any random value if no weighted scores are considered in the requested calculation.
- Column 4 to last: Scoring components values. Only numeric data is allowed. Tab is employed as field separator. A sample Data input file must look like shown below when imported into Excel. The Data files used in the CompScore algorithm validation can be downloaded from our data repository.
The user can also prepare its own Data file as long as it complies with the above described format. In addition, we freely provide a python script (ScoresDecomposer.py) that is able to summarize the scoring functions components information from Dock scoring files, OEDocking scoring tables and Gold log files. For details on using this script, please refer to its help page.
Output files
Links for downloading the results are sent to the user to the e-mail address provided at the form once its jobs is finished. If a GA optimization is requested, the user will receive two links: one for a log file and a second one for the ranked molecules file. In case a Rescoring run is requested, only the latter file will be generated. The log file contains information regarding the optimization process such as the Number of scores, Constant scores, Scores removed due to correlation, GA evolution details (every 10 generations), Score components is the best solution, Performance statistics of the best rescoring model and Execution time. The log files involved in the CompScore validation using either BEDROC or EF as enrichment metrics can be downloaded from our data repository.
Submit calculation
After all parameters required for either a GA optimization or Rescoring calculation have been provided, the calculation can be submitted by pressing the Submit button. The user will be redirected to a new page where links to download the results will become available once the requested calculation finishes. Optionally, the user can bookmark this URL to check the calculation status and results at any time.
